Exploiting SUMO Fusion Technology for Enhanced Expression of Nanobodies Targeting Vascular Endothelial Growth Factor (VEGF) in Escherichia coli
2024-10-03 | volume 2 Issue 3 - Volume 2 | Research Articles | Salim AlhafyanAbstract
In recent years, nanobodies have emerged as invaluable tools in various research and therapeutic arenas owing to their compact size, robust stability, and high specificity. Notably, vascular endothelial growth factor (VEGF) is a key regulator of angiogenesis, and plays a pivotal role in cancer and other pathological conditions. Consequently, the development of nanobodies targeting VEGF has garnered substantial attention. However, conventional expression methods for nanobodies in Escherichia coli often encounter challenges such as protein misfolding, low yields, and insolubility. In response to these hurdles, small ubiquitin-related modifier (SUMO) fusion technology has emerged as a promising strategy for enhancing the soluble production of nanobodies in E. coli. Here, we report the expression, purification, and characterization of VHH (variable domain of heavy chain antibodies) in an E. coli expression system using a (SUMO) fusion partner. The fusion gene (SUMO-active site TEV-VHH) was cloned into the pET-28(a+) expression vector and transferred into E. coli strain BL21 codon+. VEvhh10 expression was optimized using 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 22 hours at 25°C in Luria Broth (LB). Protein expression was observed in soluble form on 15% SDS-PAGE. The supernatant of SUMO-VEvh10 was collected and purified by Nickel-nitrilotriacetic acid (Ni-NTA) chromatography. The biological activity of VEvhh10 was assessed by examining its binding to VEGF. The successful binding of VEvhh10 to VEGF suggests that this non-fused protein could be utilized in future therapeutic and clinical diagnostic applications.
Keywords : VHH (variable domain of heavy chain antibodies), Vascular Endothelial Growth Factor (VEGF), SUMO Fusion Technology, Soluble Production.
INTRODUCTION
Angiogenesis, the branching out of new blood vessels from pre-existing vasculature, occurs physiologically during embryogenesis, the female reproductive cycle, and wound healing (1). It is also a crucial process in a variety of pathological conditions, including tumor growth, metastasis (2, 3), ischemic diseases, diabetic retinopathy (4, 5), chronic inflammatory reactions, age-related macular degeneration, rheumatoid arthritis, and psoriasis (6). Vascular endothelial growth factor (VEGF) is the most potent and predominant regulator of angiogenesis described to date (7, 8). This angiogenesis factor can instigate numerous biological responses in endothelial cells (ECs), such as survival, proliferation, migration, and vascular permeability, as well as the production of proteases and their receptors, creating prime conditions for angiogenesis (9, 10). It is estimated that up to 60% of human cancer cells express VEGF to create the vascular network necessary to support tumor growth and metastasis. Inhibiting angiogenesis has become an intensely investigated pharmaceutical area and represents a promising strategy for the treatment of cancer and several other diseases (11). Because of its central role in pathological angiogenesis, VEGF is a major therapeutic target. Strategies aiming to block the binding of VEGF to its receptors or to block intracellular signaling events form the basis of many new developments in anti-angiogenic cancer therapy (12). Numerous substances have been developed as angiogenesis inhibitors, some of which have already been approved for clinical use. These include monoclonal anti-VEGF antibodies (bevacizumab and ranibizumab) (13), anti-VEGF aptamers (pegaptanib), and VEGF receptor (VEGFR) tyrosine kinase inhibitors (sorafenib and sunitinib) (14, 15). Single-domain antibodies, also known as nanobodies or VHHs, possess valuable characteristics such as effective tissue penetration, high stability, ease of humanization, efficient expression in prokaryotic hosts, and high specificity and affinity for their respective antigens. Consequently, they can be introduced as alternative therapeutic candidates to traditional antibodies. VHHs represent the smallest functional unit of an antibody, preserving all of its functions, and due to their minimal size, they are also recognized as nanobodies (16, 17). Studies conducted by Shahngahian and colleagues in 2015 demonstrated that VEvhh10 (accession code LC010469) has a potent inhibitory effect on the binding of VEGF to its receptor (18). This VHH exerts its inhibitory role by binding to the VEGF receptor binding site. Among the members of the VHH phage display library, VEvhh10 possesses the highest binding energy at the VEGF receptor binding site, covering vital amino acids involved in the biological activity of VEGF and disrupting its function (18). The conventional expression of nanobodies in E.coli faces several challenges, primarily stemming from their small size and complex folding requirements. Nanobodies often exhibit low solubility, leading to the formation of inclusion bodies and hampering their functional utility. Moreover, the intricate disulfide bond formation and protein folding pathways of nanobodies make them prone to misfolding and aggregation within the bacterial cytoplasm (19-21). One standard method for expressing non-fused VHH is the use of E. coli expression systems. However, expressing non-fused VHH using conventional cytoplasmic expression methods in this prokaryotic system often faces challenges, including low expression, poor solubility, and misfolding of the antibody in E. coli (22-24). To overcome these shortcomings, we chose a novel expression system using a small ubiquitin-related modifier (SUMO) molecular partner (25, 26). SUMO Fusion Technology has emerged as a powerful tool for enhancing the soluble expression of proteins, including nanobodies, in E. coli. By fusing the target protein with the SUMO protein, researchers can promote proper folding, enhance solubility, and increase expression yields. SUMO fusion tags facilitate protein purification and can be cleaved post-purification to yield the desired protein product in its native form (27). SUMO is covalently attached to other proteins and plays roles in post-translational modifications (28). These roles include significantly increasing the yield of recombinant proteins, facilitating the correct folding of the target protein, and promoting protein solubility (29). The aim of this study was to develop an alternative method for more efficient production of VHH nanobodies in an E. coli-based expression system using the SUMO fusion tag. The SUMO fusion tag improves the solubility and yield of VHHs. Our results demonstrated that SUMO is very effective in promoting the soluble expression of VHH in E. coli. The resulting recombinant bioactive VHH can be used for therapeutic applications and clinical diagnosis in the future.
MATERIALS AND METHODS
Molecular and Chemical Materials
The materials, chemicals, and reagents required for the lab are listed as follows:
Ampicillin, kanamycin, and agarose from Acros (Taiwan), IPTG from SinaClon (Iran)
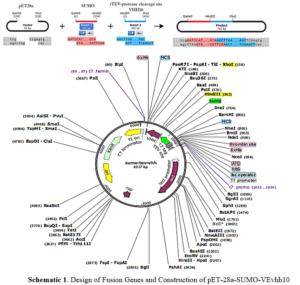
Ni-NTA resin from Qiagen (Netherlands), Plasmid extraction kit and gel extraction kit from GeneAll (South Korea), Enzyme purification kit from Yektatajhiz (Iran), Restriction enzymes HindIII/XhoI, ligase enzyme, and other molecular enzymes from Fermentas (USA), Pfu polymerase and Taq polymerase from Vivantis (South Korea), Primers from Sinagen (Iran), Methylthiazole-tetrazolium (MTT) powder from Sigma (USA), Penicillin-Streptomycin, Trypsin-EDTA, and DMEM-low glucose from Bio-Idea (Iran), Fetal Bovine Serum (FBS) from GibcoBRL (USA), Monoclonal conjugated anti-human antibody with HRP from Pishgaman Teb (Iran), Anti-Austen antibody from Roche (Switzerland), Other chemicals from Merck (Germany). Schematic 1 shows a schematic diagram of the construction of the pET-28a-SUMO-TEV-VEvhh10 Gene using SnapGene v5.1.5.
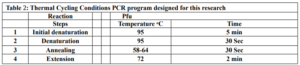
First, suitable primers were designed by OligoAnalyzer to isolate the SUMO gene sequence (see Table 1). The plasmid was used as a template for the polymerase chain reaction (PCR). Amplification was performed using the Pfu polymerase enzyme in a thermal cycler, utilizing the software we designed for this research (see Table 2). Different temperatures were tested (58, 59.5, 61, 62.5, 64) for primer annealing to the PCR template, with 58 °C being selected as the optimum temperature. The restriction enzyme sites were incorporated at the beginning and end of the primers. Additionally, the TEV protease cleavage site was placed between the SUMO and VEvhh10 sequences.
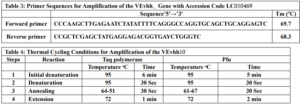
Gene Cloning: Primers for the amplification of the VEvhh10 gene with the accession code LC010469 were initially designed (Table 3). The plasmid containing the gene fragment served as a template for the PCR reaction. Amplification was carried out using Taq polymerase and Pfu polymerase enzymes in a thermocycler with a programmed temperature profile (Table 4). Various annealing temperatures for primer binding to the template were tested, and a temperature of 65°C was found to be the optimal annealing temperature. Primer cutting sites at the beginning and end of the Gene were designed, and the location of the TEV protease enzyme cutting site was positioned between the SUMO and the VEvhh10 sequence.
The gene fragments of SUMO and VEvhh10, amplified using Pfu polymerase via PCR, were extracted from the gel. Simultaneously, the amplified fragments and the pET-28a vector were digested using the BamHI and XhoI restriction enzymes. Purification was carried out using a commercial enzyme purification by GeneAll kit. Subsequently, the gene fragments were ligated into the target vector using T4 DNA ligase enzyme. The resulting ligated product was incubated overnight at 4°C. The ligated product was then transformed into E. coli DH-5α bacteria. To confirm the insertion of the Gene into the vector, the transformed bacteria were plated on kanamycin antibiotic-containing LB agar plates. Finally, the obtained colonies underwent Colony PCR using specific primers targeting VHH, T7 promoter, and terminator regions. The PCR products were analyzed on a 1% agarose gel, and positive transformants were screened and cultured further. The non-recombinant plasmid was purified using a plasmid extraction kit, and enzymatic digestion was performed to confirm the insertion of the gene fragment into the plasmid.
Expression and Soluble Detection of Recombinant SUMO-VHH
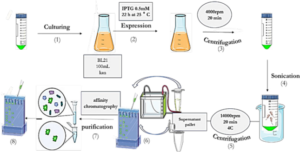
As depicted in Schematic 2, the pET-28a plasmid containing SUMO and VHH was transformed into E.coli BL21 (DE3) expression host cells using the heat shock method. The accuracy of transduction was confirmed by isolating colonies grown on LB agar medium supplemented with 50 mg/mL kanamycin. To express the genotype protein, a colony of bacteria containing the recombinant expression plasmid was inoculated into 10 mL of LB culture medium supplemented with 50 mg/mL kanamycin antibiotic and incubated at 37°C with optimal aeration at 250 rpm. Subsequently, 1 mL of mature bacteria was transferred to 50 mL of LB medium containing kanamycin antibiotics and incubated at 37°C until the OD600 reached approximately 0.6. Expression was induced by adding 0.5 mM IPTG and continued for 22 hours at 25°C. Following expression, the resulting product was centrifuged at 4000 rpm for 15 minutes at 4°C, and the bacterial cells were sonicated in lysis buffer (pH 8). The sonication product was centrifuged again at 12,000 rpm for 20 minutes at 4°C, and the supernatant was analyzed using the SDS-PAGE method (30). Proteins were purified by gradient chromatography using a nickel agarose column. The protein sample was transferred to the column pre-equilibrated with wash buffer (50 mM Tris-base, pH 8.0, 300 mM NaCl, 20 mM imidazole). Proteins that did not bind to the column due to the lack of histidine sequence were removed. Only the target protein remained attached to the column due to the presence of the histidine sequence. The bound proteins were eluted using elution buffer (50 mM Tris-base, pH 8.0, 300 mM NaCl, 250 mM imidazole). Purification was conducted using cold buffers to prevent thermal degradation of the proteins. Protein concentration was determined using the Bradford method with BSA as the protein standard (31).
The expression and purification of VEGF8-109
The His-tagged VEGF8-109-RBD was expressed in E.coli BL21 (DE3) bacterial cells and subsequently purified using a Ni-NTA agarose column, following previously described protocols with slight modifications (32). Protein expression and purification were analyzed through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue R250 staining. To remove excess salt, the purified protein was dialyzed three times against phosphate-buffered saline (PBS) containing 10% (v/v) glycerol at 4 °C for 12 h. Protein concentration was estimated using the Bradford assay, with BSA as a standard (31).
ELISA-Based Immunoassay
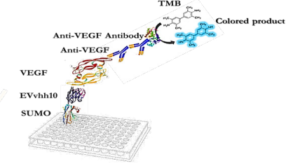
The designed immunoassay is shown in Schematic 3. First, 100 μL of SUMO-VHH recombinant protein in carbonate-bicarbonate buffer was added to each well of a cell ELISA and incubated for 16 hours at room temperature. After 16 hours, the supernatant solution was dried and washed three times with 100 μL of PBS buffer. The blocking step was performed using a PBS solution containing 2% gelatin in a volume of 350 μL and placed at 37°C for 1 hour. Subsequently, the blocking buffer was dried and washed three times with 100 μL of PBST buffer (PBS + 0.05% Tween-20). Next, 100 μL of serially diluted VEGF solutions (ranging from 0.5 ng/mL to 1000 ng/mL) were added to the wells and incubated for 2 hours at room temperature. After incubation, all wells were dried and thoroughly washed three times with PBST buffer. Following this, 100 μL of human anti-VEGF monoclonal antibody at a concentration of 1000 ng/mL was added to the wells. After washing with PBST, 100 μL of HRP-conjugated anti-human IgG antibody was added to each well and incubated in the dark at 37°C for 1.5 hours. Subsequently, 100 μL of TMB was added to each well and incubated for 15 minutes in the dark. Finally, 100 μL of 2 N sulfuric acid was added to each well, and the absorbance of the wells was read at a wavelength of 450 nm (33).
RESULTS
Products of the PCR Reaction
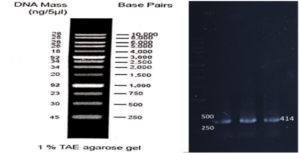
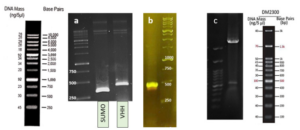
Based on our previous study (34), the VEvhh10 gene was amplified using pfu polymerase at an annealing temperature of 65 °C as showed in (Fig. 1).
Construction of pET28a-SUMO-VHH Expression Plasmid
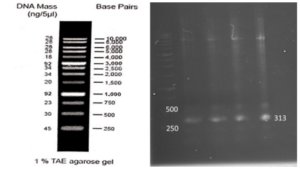
As depicted in (Fig. 3), the VEvhh10 product (402 bp) was amplified and double digested using HindIII and XhoI enzymes. The resulting product underwent purification in the preceding steps. The SUMO product, generated by the Pfu enzyme, was double digested using HindIII and BamHI enzymes, and the product (304 bp) was purified. Additionally, the plasmid pET28a was double digested using BamHI and XhoI enzymes, and the resulting product was also purified in preparation for the conjugation process.
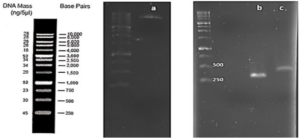
After joining the aforementioned sequences with the presence of the T4 DNA ligase enzyme, the respective products were incubated at 4°C for 14 hours and transformed into E. coli DH5α. The transformed product was cultured on a kanamycin plate, as shown in (Fig. 4.b). Agar plates containing the antibiotic ampicillin were labeled as a negative control, as depicted in (Fig. 4.a). Subsequently, in (Fig 4. c), some samples were isolated and confirmed using colony PCR by specific primers related to the T7 Promoter and Terminator, as shown in (Fig 4.d), and specific primers related to VHH, as depicted in (Fig. 4.e).
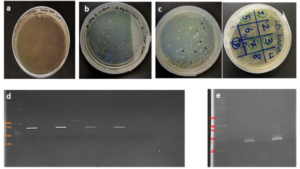
Confirmation of Gene Cloning
A fresh single colony was grown in LB agar containing the antibiotic kanamycin, and the extracted plasmid was used as a template to amplify the target gene and construct the SUMO-VHH. This step aimed to confirm the correctness of the gene cloning stage. PCR was carried out using the Forward and Reverse primers of the VEvhh10 gene (414 bp) and SUMO gene (approximately 310 bp), as depicted in (Fig. 5.a), as well as the Forward T7 promoter Reverse SUMO primers (approximately 470 bp), as shown in (Fig. 5.b). To ensure further reassurance, the plasmid was double digested using the cutting enzymes BamHI and XhoI. The double enzymatic digestion process was performed, and the desired Gene was observed on the gel, as illustrated in (Fig. 5.c). The results indicated the success and confirmation of gene cloning.
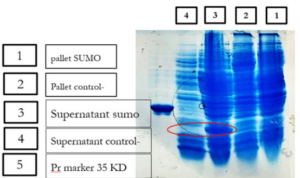
Expression and Purification of the SUMO_VHH
E.coli cells harboring SUMO-VHH were induced by 0.5 mM IPTG for 22 h at 25°C. The cell pellets were harvested by centrifugation, and protein was extracted and separated by sonication and centrifugation. The supernatants and precipitate were collected and subjected to 12% SDS-PAGE analysis. As depicted in (Fig. 6), the expression of a 24 kDa protein, similar to the predicted size, was induced by IPTG, compared with the negative control (blank plasmid) or recombinant bacteria without IPTG induction.
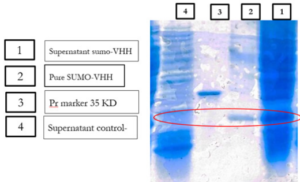
Previous studies have indicated that recombinant antibodies often form inclusion bodies using traditional E. coli expression methods (35). Our results suggest that SUMO is very helpful in promoting the soluble expression of VHH (36). To obtain high-purity recombinant protein, Ni-NTA chromatography was employed for purification because the 6His-tag was located in the N-terminal of SUMO. Different concentrations of imidazole were tested to elute the recombinant protein. The results showed that SUMO-VHH was efficiently eluted from the Ni-NTA column using an elution buffer containing 250 mM imidazole. SDS-PAGE analysis demonstrated that the purity of SUMO-VHH exceeded 90%, as shown in (Fig. 7).
Expression and Purification of Recombinant VEGF 8-109
As indicated by our previous research, recombinant VEGFRBD8-109 was produced in soluble form in E. coli BL21 (DE3) strain after induction for 22 hours at 24°C. Purification of the His-tagged fusion protein was performed using Ni-NTA affinity chromatography. The activity of the unconjugated VEGF RBD 8-109 produced was also evaluated through its effect on the growth and proliferation of human umbilical vein endothelial cells (HUVECs) using the MTT assay (37, 38). Cell proliferation was good with an increase in the concentration of unfused VEGF RBD 8-109. At a concentration of 240 ng/ml, the cell count reached approximately 80% compared to the sample lacking VEGF8-109 (34). Accordingly, it can be used in VHH-SUMO binding assays.
Investigating the Binding Ability of Recombinant SUMO-VEvhh10 with VEGF RBD 8-109
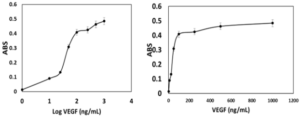
To verify the binding ability of recombinant SUMO-VEvhh10 to VEGF, an ELISA-based immunoassay was utilized according to the method described earlier. The results of the dose-response curve of VEvhh10-SUMO expressed using the ELISA method (Fig. 8) demonstrated that as the VEGF concentration increased, more anti-VEGF molecules were attached, resulting in increased light absorption.
DISCUSSION
The success of this study in amplifying, cloning, and expressing the VEvhh10 and SUMO genes underscores the utility of pfu polymerase and enzyme-based digestion for accurate gene construction as depicted in (Fig. 1), (Fig. 2), (Fig. 3) and (Fig. 5). The transformation of the ligated products into E. coli DH5α and subsequent colony PCR verification demonstrate that this method is efficient for cloning single-domain antibodies like VHH. The ability to amplify and clone the VEvhh10 gene with precision using pfu polymerase is consistent with earlier reports, which highlight the enzyme’s high fidelity in DNA replication and amplification compared to Taq polymerase. This methodological choice was critical in ensuring the accuracy of the VEvhh10 sequence during amplification (34). Moreover, the fusion of SUMO to VHH played a crucial role in enhancing the soluble expression of VHH. Previous studies have shown that recombinant VHHs often suffer from poor solubility and tend to aggregate into inclusion bodies when expressed in E. coli (35). The presence of SUMO, a small ubiquitin-like modifier, aids in overcoming this limitation by acting as a solubility-enhancing partner. SUMO fusions not only improve the solubility of the target nanobodies but also stabilize it during expression in bacterial hosts (36). In this study, SUMO fusion resulted in high yields of soluble VHH, which was efficiently expressed and purified using Ni-NTA chromatography with purity exceeding 90% as depicted in (Fig. 6) and (Fig. 7). This high purity, confirmed by SDS-PAGE, highlights the effectiveness of SUMO in both solubility enhancement and streamlined purification using His-tag. The successful production of recombinant VEGF 8-109 and its biological activity in promoting HUVEC proliferation further supports its use in functional assays. The MTT assay results, showing increased cell proliferation in response to VEGF 8-109, are consistent with previous research that indicates the importance of VEGF in promoting angiogenesis and endothelial cell growth (34). This validates the functional integrity of the expressed VEGF 8-109, suggesting that it retains its biological activity post-purification. The recombinant VEGF 8-109 can now serve as a valuable reagent in future binding assays with VHH or other therapeutic antibodies targeting VEGF pathways. Furthermore, the ELISA-based binding assays confirmed that SUMO-VEvhh10 binds effectively to VEGF, with increasing concentrations of VEGF resulting in a corresponding increase in light absorption as depicted in (Fig. 8). This binding affinity is crucial, especially for applications where VHH antibodies might be used to inhibit VEGF-mediated signaling, which is implicated in pathological conditions such as cancer and age-related macular degeneration (39). The observed binding behavior is consistent with previous works where VHH antibodies, despite their small size, exhibit strong binding capabilities to their respective antigens. This reinforces the potential of SUMO-VEvhh10 as a therapeutic agent targeting VEGF-related diseases, and the use of the SUMO fusion strategy could offer advantages over traditional expression systems by improving stability, solubility, and yield. Comparatively, the SUMO fusion system has demonstrated its superiority over other solubility-enhancing tags such as GST or MBP in maintaining the bioactivity of recombinant proteins (40, 41). While GST and MBP are effective in promoting solubility, they can sometimes mask epitopes or interfere with downstream applications, making SUMO a preferable option in cases where antigen binding and bioactivity must be preserved (42). This study adds to the growing body of literature that underscores the effectiveness of SUMO fusion in producing functional and soluble VHH antibodies for therapeutic applications. Future work could focus on optimizing expression conditions to further enhance yield or test the efficacy of SUMO-VEvhh10 in animal models of VEGF-related diseases.
CONCLUSION
This study highlights the utility of SUMO Fusion Technology for enhancing the expression of VEvhh10 targeting VEGF in E. coli. The successful production of soluble VHH against VEGF underscores its potential for future therapeutic and diagnostic applications in various diseases.
References :1. Dudley AC, Griffioen AW. Pathological angiogenesis: mechanisms and therapeutic strategies. Angiogenesis. 2023;26(3):313-47.
2. Ma Q, Reiter RJ, Chen Y. Role of melatonin in controlling angiogenesis under physiological and pathological conditions. Angiogenesis. 2020;23(2):91-104.
3. Tan S, Zang G, Wang Y, Sun Z, Li Y, Lu C, Wang Z. Differences of angiogenesis factors in tumor and diabetes mellitus. Diabetes, Metabolic Syndrome and Obesity. 2021:3375-88.
4. Jo DH, Kim JH. Toward the clinical application of therapeutic angiogenesis against pediatric ischemic retinopathy. Journal of Lipid and Atherosclerosis. 2020;9(2):268.
5. Wu W, Tang L. Research progress of anti-angiogenesis drug targets in diabetic retinopathy. [Zhonghua yan ke za Zhi] Chinese Journal of Ophthalmology. 2014;50(11):871- 6.
6. Kelly AG, Panigrahy D. Targeting angiogenesis via resolution of inflammation. Cold Spring Harbor Perspectives in Medicine. 2023;13(3):a041172.
7. Gómez-Bernal F, Fernández-Cladera Y, Quevedo-Abeledo JC, García-González M, González-Rivero AF, de Vera-González A, et al. Vascular endothelial growth factor and its soluble receptor in systemic lupus erythematosus patients. Biomolecules. 2022;12(12):1884.
8. Alewine C. Macrophages Under the Influence of Tumor Mesothelin Weaken Host Defenses against Pancreatic Cancer Metastasis. Cancer Research. 2024;84(4):513-4.
9. Miller B, Sewell-Loftin MK. Mechanoregulation of vascular endothelial growth factor receptor 2 in angiogenesis. Frontiers in Cardiovascular Medicine. 2022;8:804934.
10. Lee J-H, Parthiban P, Jin G-Z, Knowles JC, Kim H-W. Materials roles for promoting angiogenesis in tissue regeneration. Progress in Materials Science. 2021;117:100732.
11. Melincovici CS, Boşca AB, Şuşman S, Mărginean M, Mihu C, Istrate M, et al. Vascular endothelial growth factor (VEGF)-key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59(2):455-67.
12. Zhao M, Guan P, Xu S, Lu H, Liu Z. Molecularly imprinted nanomedicine for anti- angiogenic cancer therapy via blocking vascular endothelial growth factor signaling. Nano letters. 2023;23(18):8674-82.
13. Sharma A, Parachuri N, Kumar N, Sharma R, Bandello F, Kuppermann BD, Loewenstein A. Brolucizumab—another anti-VEGF or beyond. Eye. 2020;34(9):1499-500.
14. Riccardi C, Napolitano E, Platella C, Musumeci D, Melone MA, Montesarchio D. Anti‐VEGF DNA‐based aptamers in cancer therapeutics and diagnostics. Medicinal Research Reviews. 2021;41(1):464-506.
15. Das A, Mahapatra S, Bandyopadhyay D, Samanta S, Chakraborty S, Philpotts LL, et al. Bleeding with vascular endothelial growth factor tyrosine kinase inhibitor: a network meta- analysis. Critical Reviews in Oncology/Hematology. 2021;157:103186.
16. Jin B-k, Odongo S, Radwanska M, Magez S. Nanobodies: a review of generation, diagnostics and therapeutics. International journal of molecular sciences. 2023;24(6):5994.
17. Zhang Q, Zhang N, Xiao H, Wang C, He L. Small Antibodies with Big Applications: Nanobody-Based Cancer Diagnostics and Therapeutics. Cancers. 2023;15(23):5639.
18. Shahangian SS, Sajedi RH, Hasannia S, Jalili S, Mohammadi M, Taghdir M, et al. A conformation-based phage-display panning to screen neutralizing anti-VEGF VHHs with VEGFR2 mimicry behavior. International journal of biological macromolecules. 2015;77:222- 34.
19. Li W. Distinct enzymatic strategies for de novo generation of disulfide bonds in membranes. Critical Reviews in Biochemistry and Molecular Biology. 2023;58(1):36-49.
20. Cao W, Wang H, Quan M, Li Y, Su Y, Li Y, et al. Reversible control of tetrazine bioorthogonal reactivity by naphthotube-mediated host-guest recognition. Chem. 2023;9(10):2881-901.
21. Campbell E, Luxton T, Kohl D, Goodchild SA, Walti C, Jeuken LJ. Chimeric Protein Switch Biosensors. Springer; 2024.
22. Bu D, Zhou Y, Tang J, Jing F, Zhang W. Expression and purification of a novel therapeutic single-chain variable fragment antibody against BNP from inclusion bodies of Escherichia coli. Protein expression and purification. 2013;92(2):203-7.
23. Yuasa N, Koyama T, Fujita-Yamaguchi Y. Purification and refolding of anti-T-antigen single chain antibodies (scFvs) expressed in Escherichia coli as inclusion bodies. BioScience Trends. 2014;8(1):24-31.
24. Landeta C, Boyd D, Beckwith J. Disulfide bond formation in prokaryotes. Nature microbiology. 2018;3(3):270-80.
25. Sumikawa T, Nakakido M, Matsunaga R, Kuroda D, Nagatoishi S, Tsumoto K. Generation of antibodies to an extracellular region of the transporters Glut1/Glut4 by immunization with a designed antigen. Journal of Biological Chemistry. 2024;300(2).
26. Huang Z, Hua H, Du X, Zhen Z, Zhao W, Feng J, Li J-a. A specific nanobody-based affinity chromatography resin as a platform for small ubiquitin-related modifier fusion protein purification. Journal of Chromatography A. 2024;1713:464508.
27. Allen MC, Karplus PA, Mehl RA, Cooley RB. Genetic Encoding of Phosphorylated Amino Acids into Proteins. Chemical Reviews. 2024.
28. Chato-Astrain I, Pronot M, Coppola T, Martin S. Molecular Organization and Regulation of the Mammalian Synapse by the Post-Translational Modification SUMOylation. Cells. 2024;13(5):420.
29. Zhang M, Zheng Y, Wang S, Wang P, Huang J, Song X, et al. Soluble expression of recombinant human interleukin-2 in Escherichia coli and its facile production. Protein Expression and Purification. 2024:106507.
30. Harithpriya K, Ganesh GV, Umapathy D, Ramkumar K. SDS-PAGE Analysis. Advanced Mammalian Cell Culture Techniques: CRC Press; 2024. p. 66-9.
31. Nouroozi RV, Noroozi MV, Ahmadizadeh M. Determination of protein concentration using Bradford microplate protein quantification assay. Disease and Diagnosis. 2015;4(1):11-7.
32. Rezaei S, Takalloo Z, Rezaei ZS, Babaeipour V, Talebi AF, Sajedi RH. Soluble overexpression, high-level production and purification of receptor binding domain of human
VEGF8-109 in E. coli. Process Biochemistry. 2020;96:228-38.
33. Tabatabaei MS, Ahmed M. Enzyme-linked immunosorbent assay (ELISA). Cancer cell biology: methods and protocols: Springer; 2022. p. 115-34.
34. Salim A. Molecular Cloning, Cell-surface displayed expression of VHH against VEGF Expressed in E. coli by ice nucleation protein (INP). SJSI. 2024;2-2.
35. Shibaei N, Majidi J, Bashir NS, Karkhaneh A, Razavi K. Production and partial purification of the Grapevine fanleaf virus coat protein 42 polyclonal antibody against inclusion body expressed in Escherichia coli. Iranian Journal of Biotechnology. 2018;16(4).
36. Patakottu BKR, Vedire VR, Reddy CR. Robust production of active Ulp1 (SUMO protease) from inclusion bodies. Protein Expression and Purification. 2023;211:106328.
37. Drevs J. VEGF and angiogenesis: implications for breast cancer therapy. European Journal of Cancer Supplements. 2008;6(6):7-13.
38. Laborda-Illanes A, Sánchez-Alcoholado L, Castellano-Castillo D, Boutriq S, Plaza- Andrades I, Aranega-Martín L, et al. Development of in vitro and in vivo tools to evaluate the antiangiogenic potential of melatonin to neutralize the angiogenic effects of VEGF and breast cancer cells: CAM assay and 3D endothelial cell spheroids. Biomedicine & Pharmacotherapy. 2023;157:114041.
39. García-Quintanilla L, Luaces-Rodríguez A, Gil-Martínez M, Mondelo-García C, Maroñas O, Mangas-Sanjuan V, et al. Pharmacokinetics of intravitreal anti-VEGF drugs in age-related macular degeneration. Pharmaceutics. 2019;11(8):365.
40. Loughran ST, Walls D. Tagging Recombinant Proteins to Enhance Solubility and Aid Purification. Protein Chromatography: Methods and Protocols: Springer; 2023. p. 97-123.
41. Goksu Kaya-Ozsan A, Filiz Oner A. rDNA Mediated Bioconjugates: Fusion Proteins and their Intended Use in Medicine. Current Topics in Medicinal Chemistry.
2017;17(13):1529-41.
42. Sandomenico A, Sivaccumar JP, Ruvo M. Evolution of Escherichia coli expression system in producing antibody recombinant fragments. International Journal of Molecular
Sciences. 2020;21(17):6324.
The authors declare that they have no competing interests.
Data and materials availability: All data are available in the main text or the supplementary materials.
(ISSN - Online)
2959-8591