Simulating The Effect Of The Mechanical Behavior Of The Crankshaft In Internal Combustion Engines Under The Influence Of A Range Of Materials


INTRODUCTION
Our modern age is characterized by rapid and tremendous development in science and technology. Each learner has a smartphone and accessing the internet has become a daily need, a habit for some of us, and a source of income for others. And the development of (5G) networks that revolutionized technology and social networking for people and devices “Internet of Things”. This has led to the imposition of modern requirements to prepare the individual to keep pace with the developments of this era in all the fields related to our lives. One of the most important fields is education, especially in mathematics because of its importance in the different fields of life, computer science and especially algorithms. With the advent of technology, mathematical technologies appeared in education and proved their feasibility; the use of technological innovations in teaching math prepares learners for a High-Tech centric world and develops higher mental cognitive skills, such as problem-solving, thinking, data collecting, analysis and proof. Which fall within the scope of creativity and invention [1]. Fields of mathematical technology have diversified following the technological development of computers, mobile phones and the software used in them in addition to other technologies such as interactive whiteboards, the spread of the Internet and the educational services and platforms it provides. Mathematicians were able to use all these technologies in teaching mathematics. The benefits of mathematical technology are not just for students, it has an impact on teachers as it supports the creativity of teachers as learners and task designers and provides the opportunity to develop many new mathematical meanings [2]. Several scholars have investigated the technological pedagogical content knowledge (TPACK) for math teachers. Alternatively, a subset of them: Mailizar and Fan (2019) investigated Indonesian math teachers’ technological pedagogical content knowledge. The study used a questionnaire, and the sample consisted of (341) math teachers. The results showed that the understanding of mathematical technology ranked low and suggested more training courses for teachers [3]. In Malaysia, Bakar, Maat and Rosli’s (2020) study aimed to determine the math teacher’s self-efficacy in integrating technology and (TPACK). The study used a questionnaire containing (71) items, and the sample consisted of (66) national secondary math teachers. The results showed no gender or educational experience differences [4]. In Kenya, Mukenya, Martin and Shikuku (2020) investigated the knowledge and skills of math teachers to integrate ICT into secondary school education. The study used a questionnaire, and the sample consisted of (218) math teachers and heads of departments. The results indicated that teachers need more knowledge and skills to use ICT. They suggested that the Ministry of Education should work on policies to develop teachers’ ICT pedagogy and review the curriculum [5]. In Spain, the study of Gómez-García, Hossein-Mohand, Trujillo-Torres and Hossein-Mohand (2020) investigated the training and use of ICT in teaching mathematical concepts. The study used a questionnaire, and the sample consisted of (73) high school math teachers. The results showed differences in favor of teachers with less education experience and no gender differences [6]. Spangenberg and De Freitas (2019) in South Africa investigated the levels of (TPACK) and ICT integration barriers. The study used a quantitative questionnaire, and the sample consisted of (93) math teachers. The results showed poor technological content knowledge and suggested continuous professional development programs for teachers in specific ICT integration [7]. In Turkey, the study by Ozudogru and Ozudogru (2019) investigated math teachers’ technological pedagogical content knowledge. The study used a questionnaire containing (39) items, and the sample consisted of (202) math teachers. The technological knowledge section results showed significant differences in gender in favor of males and no differences in teaching experiences or school level [8]. In addition, the study of Birgin, Uzun and Akar (2020) investigated Turkish mathematicians’ perceptions of their proficiency in using ICT in teaching. The study used a descriptive survey; the sample consisted of (242) math teachers. The results showed that teachers’ knowledge of mathematical software is low, and there are no gender differences. However, there are differences in favor of teachers with less experience in education in terms of efficiency [9]. In China, Tan and Jiang (2021) aimed at the mathematical technological knowledge of elementary school math teachers. The study adopted the qualitative paradigm and a sample of (24) math teachers. The results showed that the teacher’s knowledge and use of technology classification are good. The previous research has yet to study the relationship between teachers’ knowledge and teachers’ training courses, academic qualifications, and teachers’ Internet access. Accordingly, this study will contribute to bridging this research gap.
Technological Mathematical Knowledge (TMK)
In 1986, Shulman came out with the (Pedagogical Content Knowledge) framework, which teachers need in terms of knowledge and tools to teach specific content. He considers educational technology a tool that facilitates teaching [11]. After the advent of E-learning and E-class design, Kohler and Mishra 2006 added technology as an independent regard of knowledge and not as a helping tool for teaching; (Technology knowledge) is the knowledge of technologies involving the skills of operating and using the old and new of them [12]. Also, they define the concept of (Technological Content Knowledge) as “an understanding of how teaching and learning can change when particular technologies are used in particular ways.” [13, p 65]. Thus, Schulman’s framework was expanded to (Technological Pedagogical Content Knowledge), which aims to demonstrate the necessary competencies for teachers to integrate technology with education [12]. Koehler and Mishra (2009) have embodied the framework in the “What is TPACK” study. The framework was a schematic illustrating the intersection of the three pieces of knowledge within the framework and the new knowledge resulting from its meeting with seven pieces of knowledge. As a result of the development of educational sciences and technologies, researchers [10,14,15] customized the content in the (TPACK) framework to include only mathematical content. [14] developed the Technological Pedagogical Mathematical Knowledge (TPMK) concept. [15, p 1] used the (Mathematical Technological Knowledge) concept, which they define as a “teacher’s knowledge of the technology developed as a result of exploring mathematics with technology”. This concept has an issue because some technologies are not just mathematical like an interactive whiteboard or Google apps. Similarly, [16, p 342] used the (Technological Mathematical Knowledge) concept, which they define as “the teacher’s knowledge of technological tools that can be used to represent mathematical knowledge”. However, [3, p 5] defines the broader concept of ICT-content knowledge as “knowing how to use ICT to represent, communicate, solve and explore mathematical contents, ideas, or problems without consideration of teaching approaches”. Taking advantage of these definitions, this paper defines (Technological Mathematical Knowledge) as knowledge of educational technologies hardware- and software along with how to use them to represent, explain, solve and explore mathematical content, ideas or issues regardless of the educational pedagogy, ” how to make a circle within a triangle using GeoGebra” [16, p 2].
Educational Technologies for Mathematics
Interactive Whiteboards
An interactive whiteboard is a versatile tool that allows teachers to deliver engaging lessons using various applications and educational programs [17]. Studies show it improves students’ math achievement [18]. And can benefit displaced learners in challenging environments.
Computer Algebra Systems: One of the most prominent software applications is GeoGebra. It can solve quadratic equations by graphing and accurately representing geometric transformations, statistical representation and data analysis, providing an interactive geometric environment for learners and representing shapes with a 3D environment; meanly, learning by GeoGebra improves the geometrical abilities of students [19]. In addition to its positive impact on achievement [20], it is also one of the best technological options that enriches the quality of research and mathematical conception from different perspectives that support feedback. It also provides strategies for teachers to teach according to students’ needs and facilitates learning through virtual representations that represent reality and focus on educational benefits [21]. Thus, the use of GeoGebra has a significant impact on mathematical abilities [22]. Another example is Sketchpad which combines geometry designs with algebra and calculus, curves representing descendants, then algebraic representation such as coordinates or equations and finally, a data table representation [23]. Sketchpad shares the advantage of learning through practice and developing the learner’s ability to use these applications with GeoGebra on smartphones [24].
Coding language: Scratch, for example, is a straightforward and exciting initial learning tool for understanding basic programming principles, creating educational and recreational content, building mathematical and scientific projects and simulating and visualizing experiments. Scratch not only allows learning math in an easy, effective and exciting way, but teachers also use it to teach basic mathematical principles of arithmetic and geometry [25]. In short, scratch is superior to other programming languages by attracting children to learn programming in the future [26].
Smartphone apps: are a form of distance learning and an extension of E-learning. Teachers can provide math content and follow learners anywhere, anytime by designing high-quality digital learning objects in math. Students can also learn mathematical content according to their circumstances and needs [27]. Moreover, the smartphone was the best technology for teachers during the COVID-19 pandemic [28]. It also supports applications such as Kahoot, a free educational program that supports many languages, such as Arabic, based on the play-and-response classroom system. It also helps students learn and self-evaluate, better demonstrate what they have learned, make math more exciting and vital and increase motivation to learn [29].
Online Tools: The field of education has been revolutionized by two powerful types of tools. The first type is the learning management system, such as MOODLE, an open-source program utilized in over 235 countries to support the E-learning process. Particularly effective in math education, MOODLE encourages learners to engage in cognitive thinking skills and fosters the generation of new ideas [30]. The second type is online learning resources, including Massive Open Online Courses (MOOCs), which cater to both teachers and students. These resources that are available through platforms like Coursera, Alison, Udemy and others, offer high-quality content in various specialties such as mathematics, computer science and languages. MOOCs have proven to be an invaluable resource, helping teachers enhance their professional knowledge and enabling students to access a wide range of courses, including specific mathematics courses [31], through platforms like Coursera, EdX and others. These platforms provide videos that can effectively supplement classroom learning, allowing teachers to explain complex concepts more easily.
The war in Syria had a significant impact on the education sector; it destroyed schools and displaced students, which led teachers to adopt unconventional education methods even before the COVID-19 pandemic, which was the first real challenge to educational technologies. According to McGonigal (2005) as cited in [32, p 49]“Teachers need an activating event to expose the limitations of their current knowledge”. So, what event is more challenging than war or a pandemic?
This phenomenon raises a controversial issue; did teachers have the knowledge and skills to help them cope with this crisis? And how did their knowledge and skills develop after the crisis? The current study aims to classify the technological mathematical understanding of Syria’s math teachers and the effects of demographic variables. Consequently, two questions and five related null hypotheses were formed for demographic variables, as follows:
METHODS
Participants
The online survey was shared in a Facebook group for Syrian math teachers. The researcher used the approval of the Ethics Committee of the Ministry of Education. Data was collected in the second semester of the 2021-2022 academic year. The sample was limited to (219) teachers, as shown in Table 1.
Tools
The study used a questionnaire based on [3]. The validity of the study tool was confirmed using an independent T-test and the reliability was assessed with a Cronbach-Alpha coefficient value of 0.859. Its items were classified into two parts; the first included demographic information, including gender, academic qualification, years of experience, established courses and Internet access. Part two: aimed at Technological Mathematical Knowledge, consists of (3) items intended for knowledge of educational devices, (4) items aimed at general understanding of software, (4) items aimed at knowledge of computer mathematical software, (4) items aimed at knowledge of Smartphone tools, two items on knowledge of online tools, (7) items aimed at mathematical technology content knowledge at levels:( strongly disagree, disagree, neutral, agree, strongly agree)
Data Analysis
In this study, the researcher used SPSS for statistical analysis, including coding responses into a five-point scale, calculating averages and standard deviations, conducting T-tests for validity, gender, and internet access, applying Cronbach’s alpha for reliability, using ANOVA for comparing mean responses in the case of (Academic qualification, courses, and Years of experience), and performing Fisher’s LSD test. All hypotheses were tested at a significance level of α=0.05.
RESULTS
Technological Mathematical Knowledge (TMK) of Syrian Math Teachers
Table (2) shows that the mean score of teachers’ knowledge of hardware was (3.37), which is higher than the average. In addition, their mobile knowledge was higher than their computer and interactive whiteboard knowledge, and the mean score of teachers’ knowledge of general software was (3.14), and the table shows that knowledge of Microsoft applications was the highest with average (3.92), the average knowledge of mathematical software was (2.53) which is below average, dynamic applications such as GeoGebra appear as the highest mean (2.84), the mean score of mobile tools was (3.47), which is higher than the average, and social media apps show the highest mean score (3.84), the mean score of online tools was (2.70), but the mean score of using mathematical technology was (2.30), which is below the average, and the highest field of use was in geometry with an average of 2.41.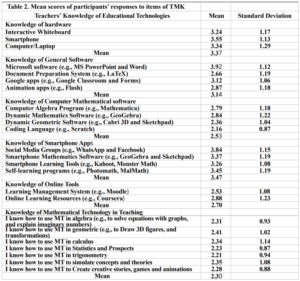
The Effects of Demographic Variables on Technological Mathematical Knowledge.
Gender differences in teachers’ (TMK)
Table 3 shows the results of an independent sample t-test comparing the means of teachers’ technological mathematical knowledge based on gender. The table shows that the mean score for male teachers is 3.13 and the mean score for female teachers is 2.75, the t-value is 3.922. A higher t-value indicates a larger difference between the means, the significance level of less than 0.05 is typically considered statistically significant. In this case, the significance level is 0.000, which is less than 0.05. Based on the t-test results, we can reject the null hypothesis that there is no difference between the means of technological mathematical knowledge scores for male and female teachers. So, there is a statistically significant difference between the means, with male teachers scoring higher on average than female teachers.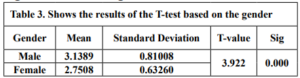
Academic qualification differences in teachers’ (TMK)
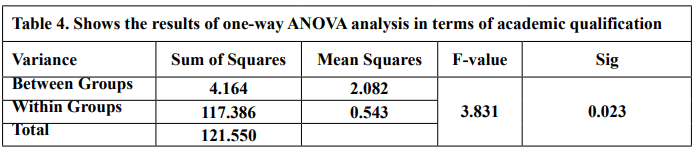
The results of the one-way ANOVA analysis in Table 4 indicate a statistically significant difference (p < 0.05) in technological Mathematical Knowledge scores between teachers with different academic qualifications. This means that we can reject the null hypothesis that there is no difference in scores between the groups.
Further analysis using the LSD test in Table 5 helps pinpoint which specific groups differ from each other. The LSD test reveals significant differences in technological proficiency scores between the following groups:
the researcher concludes that there are statistically significant differences in teachers’ (TMK) based on academic qualification in favor of the master’s degree group. At the same time, there were no differences between the bachelor and diploma groups.
Training courses differences in teachers’ (TMK)
The results of the one-way ANOVA analysis in Table 6 indicate a statistically significant difference (p < 0.05) in Technological Mathematical Knowledge scores between teachers with different training courses. This means that we can reject the null hypothesis.
The LSD test in Table 7 reveals significant differences in technological proficiency scores between the following groups:
the researcher concludes that there are statistically significant differences in teachers’ (TMK) based on Training courses in favor of the MOOCs group. At the same time, there were no differences between the No Courses and Technology Integration Courses groups.
Years of experience differences in teachers’ (TMK)
The results of the one-way ANOVA analysis in Table 8 indicate a statistically significant difference (p < 0.05) in Technological Mathematical Knowledge scores between teachers with different Years of experience. This means that we can reject the null hypothesis.
The LSD test in Table 9 reveals significant differences in technological proficiency scores between the following groups:
The researcher concludes that there are statistically significant differences in teachers’ (TMK) based on Years of experience in favor of the 1-7 years group. At the same time, there were no differences between the -14 years and 15 years and more groups.
Internet access differences in teachers’ (TMK)
Table 10 shows the results of an independent samples t-test comparing the means of teachers’ technological mathematical knowledge based on Internet access. The mean score for 3G/4G Network is 2.43 the mean score for ADSL Network is 3.85, t-value is 3.734 & (p = 0.000 < 0.05). Based on the t-test results, we can reject the null hypothesis. So, there is a statistically significant difference between the means in favor of the ADSL Network.
DISCUSSION
Results of the study showed that the general knowledge about devices was slightly above average and that a higher percentage of math teachers used smartphones because it is easy to use and widely available among learners in WhatsApp and Facebook groups as indicated in [9]. This result contradicts [3], where the highest percentage was computers. However, the researcher added an interactive whiteboard instead of the graphing calculator in our study. Our study indicates that Syrian math teachers’ general software knowledge ranked slightly above average. The highest percentage was Microsoft applications because it is familiar and easy to use and its training courses are easily accessible (ICDL). In this section, our findings are consistent with the results of [3, 9, 5], and add a section for smartphone applications, consistent with [33] in the excellent degree of using the WhatsApp application. Within the knowledge of mathematical software, the highest percentage was for GeoGebra. The reason might be to support the Arabic language and for its easy-to-use qualities. Besides, smartphone applications were more elevated than computer applications. As for Internet tools, knowledge of learning resources such as Coursera was higher than knowledge of learning management systems. This result contradicts [3] during a pre-COVID-19. This difference indicates that teachers use smartphones directly as an educational tool or a learning resource in times of crisis. The results showed poor use of mathematical technologies; a possible explanation might be that teachers are not well qualified for these technologies and not good enough at English. Another possible explanation is that most educational technological devices are unavailable in schools because the Ministry does not provide schools with such devices, which may be due to their high cost and the difficulty of producing them locally, along with the circumstances of war. This conclusion supports [5], which linked poor knowledge and use to the unavailability of technologies and devices in schools. On the other hand, [10] ranked the expertise and use of technology by Chinese math teachers as good and the integration of technology with education as excellent, owing to the availability of devices in Chinese schools.
Gender: There were significant differences in teachers’ (TMK) based on gender in favor of the male group, and this might be due to female teachers being busy with their household duties, so they do not have time to learn or use modern technological skills, unlike male teachers who have time to learn and use new technologies. This conclusion supports [8], which explains that male students tend to be more technological than female students who want to study languages and social sciences. This result is contrary to [9, 34, 6] where they showed no gender differences.
Academic qualification: There were significant differences in teachers’ (TMK) based on academic qualification in favor of the master’s degree group; a possible explanation is that master’s degree holders have excellent English and research skills. Besides, a good relationship with the Internet and all the new technologies in their specialties. As Patalinghug and Arnado [35, p 585] have pointed out “It would be a good practice for teachers to pursue advanced degrees like master’s degrees or even higher degrees” unlike the teachers who stopped at the bachelor’s or diploma, as they do not require development or scientific research. He satisfied himself with his job as a middle or secondary teacher, which does not require technical skills in our schools, [36] recommended a bachelor’s degree program should be redefined with smart technologies so students can learn fast and subjectively. Teachers might also need more time to master new technology. As [37, p 9] has mentioned, “Teachers may also feel that they do not have the time to learn new technologies because there have been many changes to middle and high school math courses and curriculum over the past several years”.
Training courses: There were significant differences in teachers’ (TMK) based on training courses in favor of the MOOCs group. This result might be because mathematical technologies are still new; therefore, they need advanced techniques that are not available in the ministerial integration courses. Logically this result supports the impact of MOOCs on teachers’ professional development and technological skills, as the studies of [38, 39] have indicated. In the USA, researchers have tested MOOCs as a teacher training course that provides content-focused experiences using technology. Expert trainers successfully designed exciting experiences for teachers that positively affected their perspectives, practices and beliefs in math teaching and statistics [39]. MOOCs worldwide allow teachers to forge partnerships and create learning communities that improve their professional knowledge and skills [40].
Years of experience: There were significant differences in teachers’ (TMK) based on years of experience in favor of the ‘1-7 years’ group. These teachers started their careers in the harshest circumstances of the war and then the COVID-19 pandemic. So, this shows that they were more resilient to learning modern technologies that helped them overcome these conditions. This result is consistent with [6], which explains that teachers with less education experience have better training in ICT and use it broadly. However, this result contradicts [8, 34], where they showed no years of experience differences.
Internet access: There were significant differences in teachers’ technological mathematical knowledge based on Internet access in favor of (ADSL); a possible explanation is that (ADSL) is more stable and cheaper in developing countries like Syria. Therefore, it allows teachers to comfortably explore the Internet, enroll in any course, such as a course on Coursera, and watch a large number of instructional videos on YouTube, unlike the limited access (3G/4G).
CONCLUSION AND RECOMMENDATIONS
The present research aimed to classify the technological mathematical knowledge of Syrian math teachers. The results showed that its classification is below average, with the highest percentage of smartphones and their mathematical applications. In the face of unprecedented challenges like war and pandemics, teachers must remain committed to developing themselves and their skills. Our research reveals a powerful tool for overcoming these obstacles: a strong relationship with the internet. By leveraging the vast resources available online, teachers can advance their mathematical and technological knowledge and equip themselves to better serve their students. This is a critical time for educators to embrace the power of technology and chart a path forward to a brighter future. This paper suggests that Ministries of Education develop comprehensive teacher training programs to prepare teachers for crises like war or pandemics. These programs should focus on developing teachers’ skills in modern mathematical software tools, mathematical applications, social media platforms, distance learning platforms, interactive lessons and E-testing. They can be extended to cover other educational subjects and mathematical technologies should be introduced to build the technological mathematical knowledge of graduates. Finally, teachers’ access to the Internet must be supported. These measures will ensure quality education during crises.

For many years, scientists have been developing ways to create biological computers using brain like tissue, or brain organoids, grown in the laboratory and connected to computer chips. The ultimate goal is to create a type of hybrid intelligence, a potentially conscious entity capable of harnessing the strengths of both the human brain and artificial intelligence. Recently, they were able to connect organisms to computer chips in a meaningful way. In 2013, scientists grew the first mini-brain in a test tube, and since then, more research has combined these lab-grown brains with electronics. “Brain-computer interface on a chip is a technology that uses a laboratory-grown “brain” (such as brain organoids) coupled to an electrode chip to achieve information interaction with the outside world through encoding, decoding, and stimulus feedback. Although artificial brains capable of walking and talking are still far in the future, brain organoids will likely be a blessing to those with neurological conditions. Similar to how other brain-based interfaces (such as Neuralink’s brain-computer interface) aim to improve the lives of individuals with neurological disorders, it is also possible to graft these brain organoids onto living tissue in the brain to stimulate neuronal growth. So, while the debate still rages over whether the future will be built with human creativity or artificial intelligence, scientists are bringing these two worlds of intelligence closer than ever before.
Researchers at the University of Colorado Anschutz Medical Campus have discovered that scents stimulate specific cells in the brain and may play a role in rapid decision-making. Researchers have discovered a new function of the hippocampus in decision- making, showing that certain cells in the brain, known as 'time cells', are stimulated by odors to facilitate quick decision-making. By tracking the activation of these cells in response to odors. It's found that smell is a stimulus that is transmitted through the nose to send nerve signals to the olfactory bulb and to the hippocampus. The two devices are closely linked. Information is processed quickly and the brain makes a decision based on the input. The team revealed a direct link between odor, hippocampal function, and associative learning, suggesting that these cells play a critical role beyond memory retrieval, and directly influence decision-making in the brain. This study shows how rats learned to associate fruity odors with reward, resulting in faster and more efficient decision-making. The scientists focused on the hippocampus, an area of the brain important for memory and learning. They knew that so-called “time cells” played a key role in hippocampal function, but they did not know their role in associative learning. The hippocampus turns on time cells to predict a decision, which would give you a glimpse into what you should remember.” “In the past, it was thought that time cells only remind you of events and time, and here we see the memory is encoded in neurons and then immediately retrieved when a decision is made.”


The national science, technology and innovation STI system in Syria consists of several universities and research centers in addition to technological productive and service, and intermediate and supporting institutions. However, this system suffers from weak relationship, interconnection, cooperation and coordination among its components for various reasons, assuring the need for building solid structures for technology transfer.
On the other hand, there is no doubt that one of the initial and necessary steps in order to invest in scientific research outputs is to protect their intellectual property (IP). In fact, there is no unified system in Syria for protecting IPs, and official responsibility for it is divided between two entities; the Directorate for Commercial and Industrial Property Protection, which is affiliated with the Ministry of Internal Trade and Consumer Protection, and the Copyright Protection Department, which is affiliated with the Ministry of Culture. Nevertheless, this role is absent at universities and other research centers, where there is no formal intellectual property policies. Hence, there is an eminent need to strengthen the ecosystem for IP protection, including the direct institutions responsible for IP granting, as well as the recognition and adherence of researchers and university students to the IP granting rules.
In this regard, the higher commission for scientific research HCSR has recently prepared an invaluable guide, entitled “A Guiding Procedures for Protecting and Investing in Scientific Research Outputs“, which primarily targets our researchers and graduate students at Syrian universities and research centers, to facilitate their acquaintance with the applicable and closely related laws and decisions collected in this guide, and to enhance the protection of intellectual property for their research outputs in preparation for investing in them and building a knowledge-based society.
The guide highlights the research outputs that researchers can protect, the procedures that must be followed in order to protect the intellectual property of these outputs, and some of the procedures for investing in them, relying in all of this on the laws, decisions and procedures in force in the Syrian Arab Republic. Additionally, the guide provides our distinguished researchers and graduate students at Syrian universities with an accurate and detailed description of the procedures that they must follow to protect and preserve the IPs of their research outputs, in preparation for their optimal investment, and strengthening national knowledge-based economy.
Finally, the guide has been divided into two chapters: the first is concerned with protecting the intellectual property of types of research outputs, and the second is concerned with investing in them. In this guide, we have also adopted the classification of scientific research outcomes into three types: applied idea – applied protocol – and prototype.
Note: the guide was prepared by Ms. Lamees Ismael, the executive director of the Syrian Journal for Science and innovation.


Previous studies of reinforced asphalt concrete have focused on different types of fiber such as polypropylene, polyester, carbon and glass (5-8). Polypropylene fibers provide three-dimensional reinforcement of the concrete, making it tougher and more durable (9,10). The common forms of these fibers are smooth-monofilament and have triangular shape. Polypropylene fibers are widely used as reinforcing agents in rigid and asphalt pavement (11,12). Othman (2010) investigated the effect of polypropylene application method on long-term aging of hot mix asphalt (HMA). Three different polypropylene application methods were prepared for that purpose and a constant polypropylene content of 0.7% by weight of the total mix was adapted. The first mixture was prepared using polypropylene coated aggregate. The second mixture was prepared using the traditional wet process method, where polypropylene is blended with asphalt binder at high temperature. The third mixture was prepared using the dry method where polypropylene powder was added to the mineral aggregate prior to mixing it with asphalt. Testing procedures included the Marshall tests for aged and unaged mixtures, indirect tensile strength, fracture energy, and unconfined compressive strength. This paper concluded that the inclusion of polypropylene has significantly improved indirect tensile strength, fracture energy, and unconfined compressive strength. It was also concluded that samples prepared using polypropylene coating methods displayed the highest tensile strength and fracture energy under aged and unaged conditions (13). Tapkın et al. (2009) concluded that the most suitable polypropylene fibers can be used at a dosage of 0.3% by weight of the aggregates and increased the Marshall stability values by 20% and the life of the Polypropylene fibers modified asphalt specimens under repeated creep loading at different loading patterns by 5–12 times versus control specimens. It also indicated that the addition of polypropylene fibers improves the behavior of the specimens by increasing the life of samples under repeated creep testing (14). Ahmad et al. (2015) studied the behavior of Polypropylene fibers reinforced asphalt mixtures on fatigue performance. The results from this study show that the addition of polypropylene fibers improves the behavior of the specimens by increasing the life of samples under fatigue testing according to the test results, the addition of 1.5 % of polypropylene fibers prolongs the fatigue life by 113 % in terms of number of cycles, in comparison to plain asphalt concrete beam (15). Zachariah et al. (2018) evaluated the effect of Polypropylene fibers reinforcement on bituminous concrete using brick as aggregates (first class brick and over burnt bricks). Resilient modulus tests, moisture susceptibility test, creep tests and indirect tensile strength test were performed. The Marshall test and basic property tests were used to justify the performance of polypropylene modified bituminous mixes using bricks as aggregates. This study concluded that brick aggregates can be used in asphalt concrete for using as a surface course if asphalt is modified with polypropylene fibers, and the optimum polypropylene fibers content was found to be 2% of aggregate by weight for first class bricks (where resilient modulus increased by 162%) and 4% of aggregate by weight for over burnt bricks (where resilient modulus increased by 157%). The results indicated that polypropylene fibers addition enhances the characteristics of asphalt, helps in reducing the temperature susceptibility of the mix, and fulfills the minimum requirement of tensile strength ratio TSR (16). Li et al. (2024) analyzed the viscoelastic characteristics of asphalt binders reinforced with polypropylene fibers by using dynamic shear rheological (DSR) testing. The binders reinforced with fiber showed superior resistance to high temperatures and long-term deformation while being less sensitive to temperature and having a more significant elastic characterization (17). Whereas Jalota et al. (2023) improved the moisture resistance of flexible pavements by using polypropylene fibers measuring 6 mm in length and different dosages of liquid anti-stripping agents (18). Other researches evaluated the influence of polypropylene fiber on concrete and rigid pavements (19,20), while other studies focused on hybrid reinforcement to improving performance of asphalt concrete mixtures through their reinforcement with two or more types of fibers such as: polypropylene and glass fibers (21,22), polyester and polypropylene fibers (23), glass and carbon Fibers (24), polyolefin and aramid fibers (25) and Hybrid Fiber and Nano (26).
MATERIAL AND METHODS
Asphalt binder
The Asphalt used in this study was a 60/70 penetration grade obtained from Homs Refinery Company. The Physical Characteristics of the Asphalt binder were tested according to standard specifications and are listed in Table 1.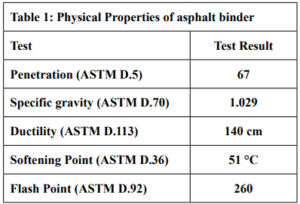 Aggregates
Aggregates
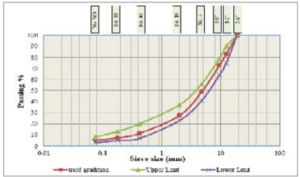
The coarse and fine aggregates were supplied from Hsia City. The gradation of the test specimens was performed in accordance with ASTM of surface course, Table 2 and Fig. 2 show the gradation of these aggregates. They were selected and incorporated in preparing all hot asphalt concrete mixes used in this study. The mechanical and physical characteristics of used aggregates have been tabulated in Table 3.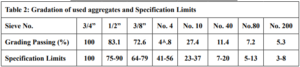
Experimental Methods
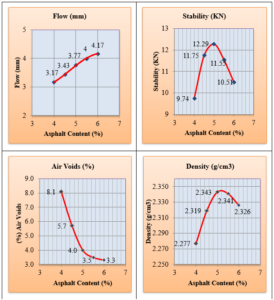
Polypropylene fibers were used in asphalt mixtures with different percentages (3, 4, 5 and 6%) by weight of the asphalt binder. polypropylene fibers were added to hot asphalt binder and mixed manually for five minutes (until the mix acquires uniformity). Then, the modified asphalt was mixed with aggregates. To determine the optimum asphalt content that would produce asphalt concrete mixtures, 15 samples were tested according to The Marshall test (ASTM.D 1559). The Marshall method is used for all mixtures. The optimum asphalt content was selected from figure. 3 as the average of the asphalt content for maximum density, maximum stability and 4% air voids. The optimum binder content was found to be 5% by weight of the total mix. All PPF modified specimens were prepared using constant asphalt binder content (5%) and produced at a mixing temperature of 160ºC.
RESULTS
Effect of Polypropylene fibers on the performance of the asphalt binder
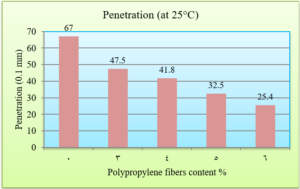
To determine the effect of Polypropylene fibers on the physical characteristics of the asphalt, penetration, softening point, and ductility tests were carried out on both reinforced and unreinforced asphalt binder with PPF. The penetration value determines the hardness of asphalt by measuring the depth (in tenths of a millimeter) to which a standard and loaded needle will vertically penetrate in 5 seconds a sample of asphalt maintained at a temperature of 25°C (ASTM D 5). The results of the penetration test are presented in Fig. 4.
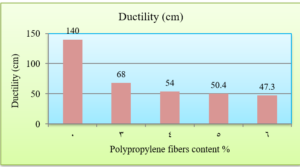
Ductility is the property of asphalt that permits it to undergo great deformation or elongation. It is defined as the distance in cm, to which a standard sample or briquette of the material will be elongated without breaking (ASTM D 113). Fig. 5 shows the change in the ductility of asphalt binder depending on the Polypropylene fiber content. As the Polypropylene content increases, the ductility values decrease.
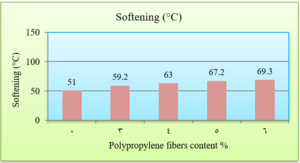
The softening point is determined as the temperature at which a sample of asphalt, subjected to a progressive increase in temperature and the weight of a steel sphere, reaches a consistence that leads to its flow through a ring of steel, until a specific deformation is obtained (ASTM D 36). The results of the softening point test are presented in Fig. 6.
Evaluation of polypropylene fibers addition on asphalt mixtures characteristics
At this stage, 45 Marshall specimens are prepared using asphalt binder content (5%). The Marshall test results for reinforced and unreinforced asphalt mixtures are tabulated in Table 5.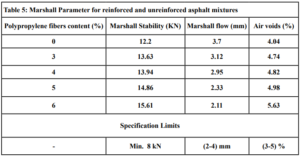
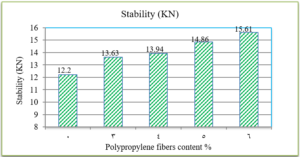
Fig. 8 indicates that as the Polypropylene fibers content increases, the Marshall flow for asphalt mixtures decreases.
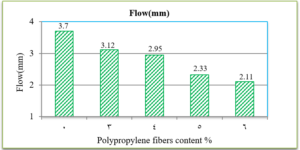
Fig. 9 presents the results of air voids percentage in the total mix for all mixtures. It indicates that polypropylene fibers have increased the air void percentage in all the specimens.
DISCUSSION
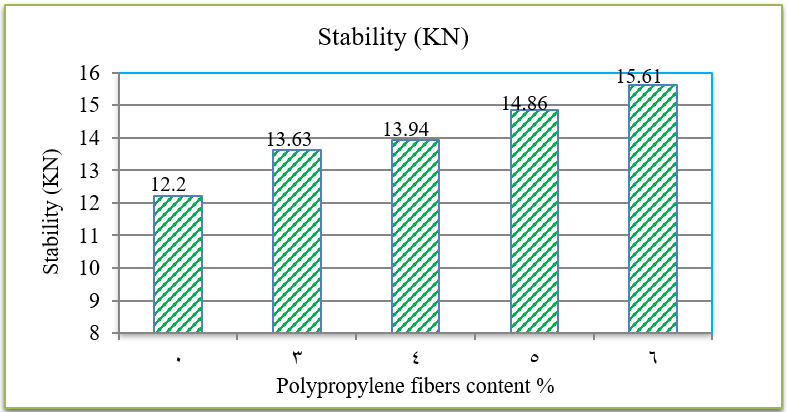
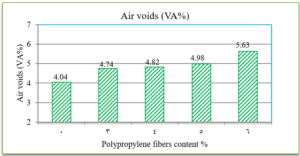
The penetration results indicate that the consistency of the PPF reinforced asphalt decreases as the PPF content in the mix increases. The reduction in penetration values were (29%, 38%, 51% and 62%) with the addition of (3%, 4%, 5% and 6%) of PPF, respectively, as compared to the unreinforced asphalt. This means that the addition of PPF makes the asphalt harder and more consistent. These results indicate that the rutting resistance of the mix is improved, but on the other hand, the high stiffness makes the asphalt concrete less resistant to fatigue cracking. It was also found in this research that the ductility values decreased as the addition percentage increased. This could be due to the position of polypropylene fibers in the cross-section of the asphalt binder during the ductility test, which prevents the asphalt from stretching easily. By increasing the Polypropylene fibers content, the softening point increased. This increase ranges from 16% to 36% with the addition of 3% to 6% of PPF content, this result indicates that the resistance of the reinforced asphalt binder to high temperatures is increased. Consequently, the results of the asphalt binder tests indicate that the reinforced asphalt samples with PPF are much stiffer and more resistant to high temperatures and rutting. The results of the Marshall tests indicate that the increasing in the polypropylene fibers content led to an increase in Marshall stability and decrease in flow values for asphalt mixtures. Increasing of 12%, 14%, 22% and 28% in stability values with the addition of 3%, 4%, 5% and 6% of PPF, respectively, to comparing with the traditional asphalt mixtures. Polypropylene fibers led to increase the air voids in all reinforced mixtures. This could have occurred because all the specimens were prepared 5% asphalt content, therefore, PPF added samples need more asphalt traditional specimens. However, the air voids of reinforced mixtures with Polypropylene fibers up to 5% content were acceptable as compared with the specification limits (3-5%) [ASTM].
CONCLUSIONS
Based on the results of studying the effect of polypropylene fibers in asphalt concrete mixtures, it can be deduced:
RECOMMENDATIONS
Journal:![]() Syrian Journal for Science and Innovation
Syrian Journal for Science and Innovation
Abbreviation: SJSI
Publisher: Higher Commission for Scientific Research
Address of Publisher: Syria – Damascus – Seven Square
ISSN – Online: 2959-8591
Publishing Frequency: Quartal
Launched Year: 2023
This journal is licensed under a: Creative Commons Attribution 4.0 International License.